Proteomic analysis of single cell clusters using laser capture microdissection
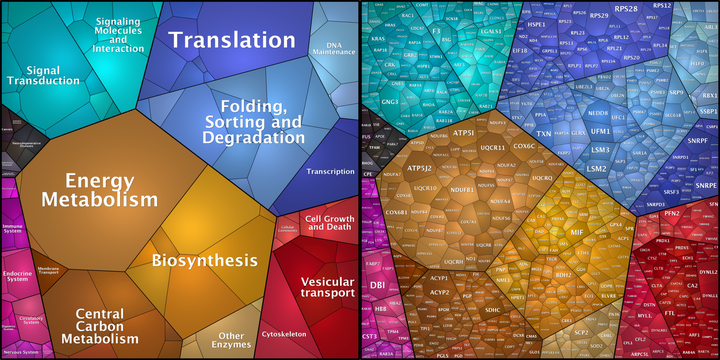 Image credit: Own Work
Image credit: Own WorkBackground
An inherent problem with the analysis of ‘bulk’ tissue by traditional proteomics methods is that the cellular heterogeneity of a tissue is often unaddressed. This heterogeneity results in averaging of the signals generated by the constituent cells and the data are dominated by those cells that are the most abundant. Isolating cells of a single type by laser capture microdissection alleviates the signal averaging caused by measuring a mixed population of cell types. Here we developed a simple, quantitative sample preparation method for sub-1000 cells isolated by laser capture microdissection.
Methods We used laser capture microdissection to isolate varying numbers of Purkinje cells from histological tissue slices of cerebellum obtained from post-mortem human donors into an adhesive coated microcentrifuge tube lid. Captured proteins were proteolytically digested with trypsin and the resulting peptides were analysed by LC-MS/MS on an Orbitrap Fusion Lumos. Data files were processed and analysed with MaxQuant and Perseus.
Preliminary Data
We were able to reproducibly detect over 2000 proteins from only 200 Purkinje cells isolated from histological tissue slices. The overlap in protein identifications was 80 % between two replicates from different tissue slices within the same donor. The proteomes generated were deep enough to detect several proteins involved in Purkinje cell development and function along with almost 600 proteins involved in general neurological cell processes. The method also shows good quantitative reproducibility between replicates (Pearson Correlation Coefficient = 0.90).
Novel Aspect
LCM-Proteomics is capable of generating reproducible, quantitative proteomes of sufficient depth from single cell types isolated from mixed populations.
Keywords
Laser capture microdissection; ultrasensitive proteomics; quantitative proteomics; neuropathology; single-cell isolation